issue contents
November 2018 issue
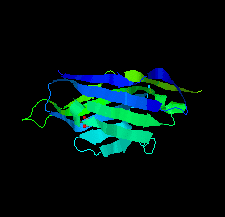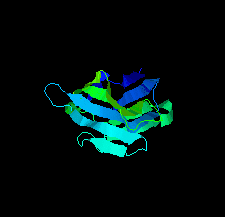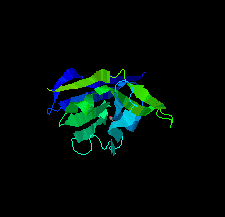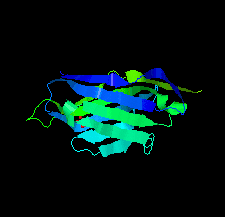
Cover illustration: A llama-derived nanobody targeting the J-base binding protein 1 [van Beusekom et al. (2018), Acta Cryst. F74, 690-695]. J-base binding protein 1 contributes to the biosynthesis and maintenance of base J, a modification of thymidine confined to some protozoa.
editorial
Free 

The Section Editor discusses the current state and future of Acta Crystallographica F and encourages readers to submit their best Research Communications.
research communications
A camelid single-domain antibody fragment (nanobody) is shown to have high affinity towards its recognition target, the J-base binding protein 1. The serendipitous crystallization of this nanobody alone, solution of its crystal structure and its refinement to 1.64 Å resolution are described. Ensemble refinement suggests that in the ligand-free state one of the complementarity-determining regions (CDRs) is flexible, while the other two adopt well defined conformations.
PDB reference: Nb6, 6fpv
Well diffracting crystals of the ectodomain of rat ectonucleotide phosphodiesterase/pyrophosphatase-3 were obtained for a construct lacking the SMB domains, while the structure of the full-length ectodomain reveals the orientation of the SMB domains but has a partially disordered active-site structure, probably owing to the absence of the active-site zinc ions.
PDB reference: ectonucleotide phosphodiesterase/pyrophosphatase-3, 6g4g
Crystal structures of the RecA-like domain 1 of Staphylococcus aureus CshA (SaCshA) and of its complex with AMP are reported; SaCshA shares a highly conserved ATP-binding mechanism with other DEAD-box RNA helicases.
BpeB, which is a major multidrug-efflux transporter in Burkholderia cenocepacia, has a C-terminal region that extends beyond those of other homologous resistance-nodulation-cell division transporters such as AcrB and MexB. Comparative modeling helped to identify truncation variants that could aid the crystallization of BpeB without eliminating its molecular functions.
Human indoleamine 2,3-dioxygenase 1 (IDO1) is a potential target for drug discovery. Three new crystal forms of human IDO1 are reported, obtained by introducing a double mutation on the surface. One of these crystal forms can diffract to high resolution and can be readily used for soaking experiments with inhibitors. This mutant represents a robust platform for determining structures of IDO1 inhibitor and substrate complexes at high resolution.
PDB references: indoleamine 2,3-dioxygenase 1, complex with epacadostat, 6e40; complex with sulfur analog of epacadostat, 6e41; complex with 4-chlorophenyl imidazole, 6e42; complex with anolog of BMS-978587, 6e43; free enzyme (ferric), 6e44; free enzyme (ferrous), 6e45; complex with tryptophan (ferrous), 6e46
Open  access
access
 access
accessStructures of N-acetyl-D-neuraminic acid lyase in the ligand-free form and of a covalent Schiff base adduct are reported. The structure and kinetic data reveal the conserved nature of these proteins among Gram-negative bacteria.
Crystal structures of (S)-3-hydroxybutyryl-CoA dehydrogenase from Clostridium acetobutylicum have been determined in apo and NAD+-bound forms. The structures, together with kinetic analyses using single-amino-acid substituted mutants, revealed the catalytically important residues in the enzyme.
Glycosyltrehalose synthase from Sulfolobus shibatae DSM5389 has an optimum catalytic cavity for the intramolecular transglycosylation mechanism in the first step of the biosynthesis of trehalose, which converts the glucosidic bond between the last two glucose residues of amylose from an α-1,4 bond to an α-1,1 bond.
PDB reference: glycosyltrehalose synthase, 5zcr
Advanced crystallogenesis methods, including microseed matrix screening, crystallization optimization by counter-diffusion and crystal detection by trace fluorescent labeling, were used in combination to successfully produce various diffracting crystal forms of a CCA-adding enzyme from the psychrophilic bacterium Planococcus halocryophilus.


 journal menu
journal menu

















![[publBio]](/logos/publbio.gif)