issue contents
July 2023 issue
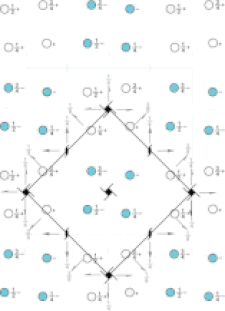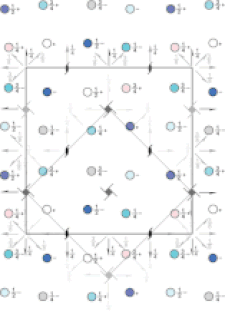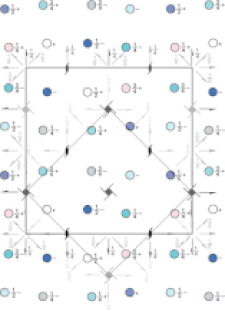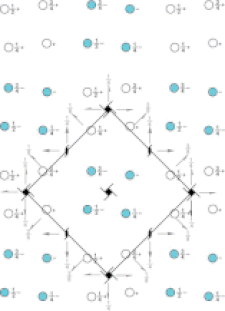
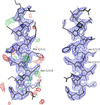
Cover illustration: Pseudo-P43212 packing of the flavin-dependent halogenase AetF [Gäfe & Niemann (2023), Acta Cryst. D79, 596–609]. Crystallographic and noncrystallographic symmetry elements are shown in black and grey, respectively. Crystallographically independent molecules are shown in different colours.
editorial
Free 

This editorial acknowledges the transformative impact of new machine-learning methods, such as the use of AlphaFold, but also makes the case for the continuing need for experimental structural biology.
CCP4
Open  access
access
 access
accessIt is shown that checkMySequence, an automated method for validating sequence assignment in cryo-EM structures of proteins, can be used to validate crystal structure models.
CCP-EM
Open  access
access
 access
accessAn integrative framework called Flexibility Hub is integrated into Scipion that simplifies the interoperability of different heterogeneity algorithms.
research papers
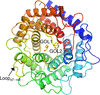
Structures of mannose 2-epimerase and a mutant from Runella slithyformis were determined in their apo form and in complex with D-glucitol. In combination with a biochemical assay, the molecular basis of the substrate specificity and activity of the enzyme was elucidated.
Open  access
access
 access
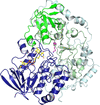
accessThe single-component flavin-dependent tryptophan halogenase AetF converts tryptophan to 5,7-dibromotryptophan during the biosynthesis of the neurotoxin aetokthonotoxin. Crystal structures of AetF with the substrates tryptophan and 5-bromotryptophan show that a flip of the indole moiety of tryptophan positions first C5 and then C7 in the same location in the active site to facilitate two successive bromination reactions.
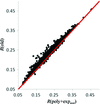
In this work, a novel method to recover the diffraction phase independently using the low-resolution data is proposed. It has been verified and found to be useful and practical using several typical examples. In addition, a practical metric has been established to determine whether the algorithm converges to a correct solution by estimating the entropy of the final results.
Open  access
access
 access
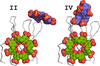
accessCalixarene-mediated protein assembly provides a basis for the development of new types of biomaterials. A fourth structure of the Ralstonia solanacearum lectin–sulfonato-calix[8]arene complex expands the crystal-engineering landscape and suggests an alternative pH trigger of assembly.
Open  access
access
 access
accessThe crystal structure of the monocupin 5-nitrosalicylate 1,2-dioxygenase, an iron(II)-dependent ring-cleaving dioxygenase, from Bradyrhizobium sp. was determined by molecular replacement using a theoretical model obtained by AlphaFold2. Comparison with structures of other members of the same class and docking of the substrate allowed identification of the residues responsible for its very unusual enzyme selectivity.
PDB reference: 5-nitrosalicylate 1,2-dioxygenase, 8ch4
Open  access
access
 access
accessThe crystal structure of a novel laccase-like multicopper oxidase from the thermophilic fungus Thermothelomyces thermophila was refined at 1.9 Å resolution. Ligand-docking simulations reveal conformational changes that influence substrate specificity.
PDB reference: laccase-like multicopper oxidase, 7zn6
Open  access
access
 access
accessThe refinement and validation of nine A-form DNA 18-mer crystal structures containing both canonical and noncanonical base pairs benefits from the use of dinucleotide conformer classes (NtCs).
addenda and errata
Open  access
access
 access
accessThe article by Afonine et al. [Acta Cryst. (2013). D69, 625–634] is corrected.

 journal menu
journal menu