issue contents
November 1998 issue
Databases for macromolecular crystallography
Proceedings of the CCP4 study weekend, January 1998
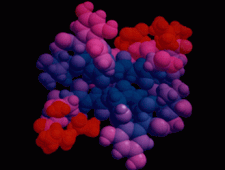
Cover illustration: The balhimycin dimer, with citrate and acetate ions in the binding pockets, colour coded on a scale where the most mobile atoms are red and the least mobile are blue. The binding pockets are clearly the most mobile, consistent with the idea that they can open and close to complex with cell-wall binding peptides. Courtesy of George Sheldrick.
research papers
Open  access
access
 access
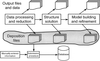
accessThe rapid growth of the World Wide Web provides major new opportunities for distributed databases, especially in macromolecular science. A new generation of technology, based on structured documents (SD) and XML, is being developed which will integrate documents and data in a seamless manner.
Open  access
access
 access
accessThe Iditis protein structure database provides the most comprehensive set of derived information about protein structure currently available and allows rapid searching for complex motifs.
Open  access
access
 access
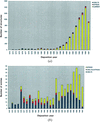
accessThe Protein Data Bank (PDB) at Brookhaven National Laboratory, a database containing experimentally determined three-dimensional structures of proteins, nucleic acids and other biological macromolecules, with approximately 8000 entries, is described.
Open  access
access
 access
accessA summary of macromlecular structure databases developed to date. The authors own work indicates that data are reported inconsistently and this should be addressed in the future.
Open  access
access
 access
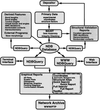
accessA description is given of how the Nucleic Acid Database (NDB) is used to study nucleic acids. In addition, the way in which the technology developed by the NDB project has been extended to macromolecules in general is summarized.
Open  access
access
 access
accessA discussion is presented of some of the issues involved in depositing and releasing macromolecular structural information, and an outline of future directions.
Open  access
access
 access
accessThe importance of validation techniques in X-ray structure determination and their relation to refinement procedures are discussed, with particular reference to atomic resolution structures. The requirements of deposition and publication, and the role of validation tools in this are analysed. The need for a rigorously defined file format is emphasized.
Open  access
access
 access
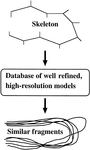
accessThe use of databases for protein crystallographic model building, refinement, validation and analysis is reviewed, and some recent developments are discussed.
Open  access
access
 access
accessA description of new analytical software tools and WWW servers for studying protein sequence, structure and function is presented.
Open  access
access
 access
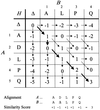
accessAlgorithms for alignment of two or more protein sequences are described. Software for sequence analysis and database searching is summarized.
Open  access
access
 access
accessThe Structural Classification of Proteins (SCOP) database is described. It provides a detailed and comprehensive description of the relationships of all known protein structures and can be used as a source of data to calibrate sequence search algorithms and for the generation of population statistics on protein structures.
Open  access
access
 access
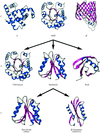
accessThe CATH database of protein domain structures classifies structures according to their (C)lass, (A)rchitecture, (T)opology or fold and (H)omologous family. Although the protocol used is mostly automatic, manual inspection is used to check assignments at some critical stages. Described in this article is a recently established facility to search the database with the coordinates of a newly determined structure.
Open  access
access
 access
accessDatabases of protein structural domains (DDBASE), aligned homologous protein structures (HOMSTRAD) and structurally aligned protein superfamilies (CAMPASS) are available on the WWW.
Open  access
access
 access
accessA survey of novel database tools for the analysis of protein–ligand interactions.
Open  access
access
 access
accessAnalysis of data from the IsoStar library shows that many hydrophobic groups exhibit strikingly strong directional preferences in their intermolecular interactions. These directional preferences may need to be taken into account in parameterizing the next generation of protein–ligand docking programs.
Open  access
access
 access
accessThe reliability and transferability of M—L bond lengths and L—M—L bond angles from crystal structure is considered in the light of the utility of tables of `typical' bond lengths in transition-metal complexes.
Open  access
access
 access
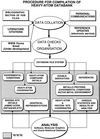
accessThe Heavy-Atom Data Bank (HAD) described contains coordinates of heavy-atom sites derived from multiple isomorphous derivatives used in protein crystallography. HAD contains information on crystallization conditions and protein binding sites that will be of value in the preparation of heavy-atom derivatives for use in preparation of isomorphous derivatives in the method of isomorphous replacement.


 journal menu
journal menu