issue contents
November 2005 issue
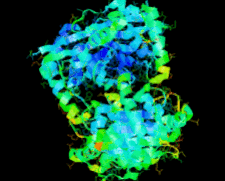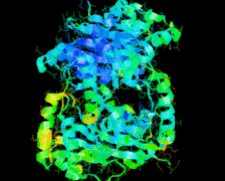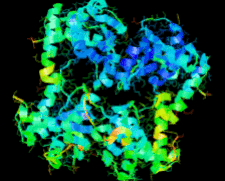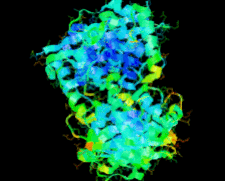
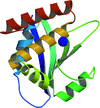
Cover illustration: The ybeY protein from Escherichia coli (p. 959).
structural genomics communications
The ybeY protein from E. coli is reported at a 2.7 Å resolution with a metal ion.
PDB reference: ybeY, 1xm5, r1xm5sf
protein structure communications
An orthorhombic crystal form of the SARS CoV main proteinase diffracting to a resolution of 1.9 Å is reported. The conformation of residues in the catalytic site indicates an active enzyme.
PDB reference: SARS coronavirus main proteinase, 2c3s, 2c3ssf
crystallization communications
Superoxide reductase is a non-haem iron-containing protein involved in resistance to oxidative stress. The oxidized form of the protein has been crystallized and its three-dimensional structure solved. A highly redundant X-ray diffraction data set was collected on a rotating-anode generator using Cu Kα X-ray radiation. Four Fe atoms were located in the asymmetric unit corresponding to four protein molecules arranged as a dimer of homodimers.
The histone chaperone cia1 from fission yeast has been overexpressed in E. coli, purified and crystallized using the vapour-diffusion method.
The nitrile hydratase from the themophilic B. smithii SC-J05-1 (Bs NHase) has been purified, cloned and crystallized.
Human Rad has been crystallized. A diffraction data set was collected to a resolution of 1.8 Å.
Further understanding of the structure and function of plasma apolipoproteins requires the determination of their high-resolution structures when complexed with lipids. In these studies, the production of homogeneous, biologically active lipoprotein particles of apolipoprotein E complexed with dipalmitoylphosphatidylcholine and their crystallization and X-ray diffraction are demonstrated.
The C-terminal domain of the transcriptional regulator Ss-LrpB from S. solfataricus was purified by affinity chromatography and crystallized. Crystals belong to space group P21212. A complete data set was collected to a resolution of 2 Å.
The carbohydrate-binding component (VP8*64–223) of the human Wa rotavirus spike protein has been overexpressed in E. coli, purified and crystallized in two different crystal forms. X-ray diffraction data have been collected that have enabled determination of the Wa VP8*64–223 structure by molecular replacement.
The isomerase domain of glucosamine-6-phosphate synthase from C. albicans has been crystallized and X-ray diffraction data have been collected. Preliminary analysis of the data reveals the oligomeric structure of the eukaryotic synthase to be a `dimer' of prokaryotic-like dimers.
The crystallization and preliminary X-ray diffraction analysis of a red marine alga lectin isolated from H. musciformis is reported.
Nucleotide-exchange factor from S. solfataricus (SsEF-1β) has been successfully crystallized. X-ray diffraction data have been collected from the native enzyme and from the selenomethionine derivative of SsEF-1β to 1.97 and 1.83 Å resolution, respectively.
Tyrosyl-tRNA synthetase from the hyperthermophilic archaeon A. pernix K1 was cloned, purified and crystallized. The crystals belonged to the tetragonal space group P43212, with unit-cell parameters a = b = 66.1, c = 196.2 Å, and diffracted to beyond 2.15 Å resolution at 100 K.
The crystallization of a hypothetical penicillin-binding protein from the archaeon P. abyssi in space group C2 by hanging-drop vapour diffusion is reported.
The mammalian Pax6 paired domain has been cocrystallizaed with a 25 bp DNA fragment of the Pax6 gene enhancer.
Recombinant rat cysteine dioxygenase (CDO) has been expressed, purified and crystallized and X-ray diffraction data have been collected to 1.5 Å resolution.
Fatty acid-CoA racemase from M. tuberculosis H37Rv has been overexpressed, purified and crystallized. Diffraction data have been collected to beyond 2.7 Å resolution using a synchrotron-radiation source.
The GluR0 ligand-binding core from N. punctiforme was expressed, purified and crystallized in the presence of L-glutamate. A diffraction data set was collected to a resolution of 2.1 Å.
Crystals of the type I Baeyer–Villiger monooxygenase (BVMO) MtmOIV from the biosynthetic pathway of mithramycin were obtained; the crystals diffracted to 2.69 Å resolution and belong to the monoclinic space group C2 (a = 143.5, b = 114.2, c = 137.8 Å β = 102.5°). Light scattering indicates that MtmOIV is a dimer of 127 kDa in solution, while in the crystalline state the data are consistent with two dimers in the asymmetric unit.


 journal menu
journal menu


























![[publBio]](http://journals.iucr.org/logos/publbio.gif)





