issue contents
December 2009 issue
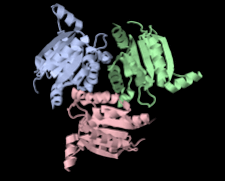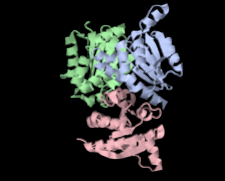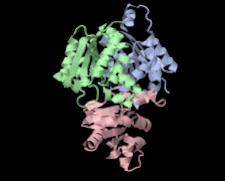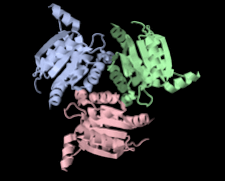
Cover illustration: Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii (Antonyuk, Strange, Ellis, Bessho, Kuramitsu, Shinkai, Yokoyama & Hasnain, p. 1200). One of a series of eight structures from RIKEN, Liverpool and Daresbury structural genomics groups which appear in this issue.
RIKEN-UK structural genomics
The structure of a protein involved in the molybdopterin and molybdenum co-factor biosynthesis pathways of Sulfolobus tokodaii has been solved to a resolution of 1.9 Å.
PDB reference: ST2315, 3iwt, r3iwtsf
The structure of the stationary phase survival protein SurE protein from the hyperthermophile Aquifex aeolicus has been solved to 1.5 Å resolution. The divalent-metal-ion-dependent phosphatase active-site pocket is occupied by sulfate ions from the crystallization medium.
PDB reference: SurE, 2wqk, r2wqksf
The structure of D-lactate dehydrogenase from Aquifex aeolicus has been determined with each subunit of the homodimer in a `closed' conformation and with the NAD+ cofactor and lactate (or pyruvate) bound at the inter-domain active-site cleft.
PDB reference: D-lactate dehydrogenase, 3kb6, r3kb6sf
The structure of ribose-5-phosphate isomerase from Methanocaldococcus jannaschii has been solved to 1.78 Å resolution, with the active site occupied by two molecules of propylene glycol mimicking the binding of a known arabinose-5-phosphate inhibitor.
PDB reference: ribose-5-phosphate isomerase, 3ixq, r3ixqsf
The structure of a putative β-phosphoglucomutase from Thermotoga maritima belonging to the haloacid dehalogenase (HAD) hydrolase family has been determined to 1.74 Å resolution.
PDB reference: TM1254, 3kbb, r3kbbsf
The crystal structure of dihydrodipicolinate synthase from the (S)-lysine synthesis pathway of Methanocaldococcus jannaschii has been solved to 2.2 Å resolution, revealing a functional homotetramer.
PDB reference: dihydrodipicolinate synthase, 2yxg, r2yxgsf
The structure of glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanocaldococcus jannaschii was determined to 1.81 Å resolution with the NADP+ cofactor at the nucleotide binding site.
PDB reference: glyceraldehyde-3-phosphate dehydrogenase, 2yyy, r2yyysf
The putative 4-amino-4-deoxychorismate lyase (TTHA0621) from T. thermophilus HB8 was cloned, overexpressed, purified and crystallized. Its crystal structure was determined by a combination of SAD and molecular-replacement methods and was refined to 1.93 Å resolution.
PDB reference: TTHA0621, 2zgi, r2zgisf
structural communications
The crystal structure of cold-shock protein E from S. typhimurium (StCspE) has been determined at 1.1 Å resolution.
PDB reference: cold-shock protein E, 3i2z, r3i2zsf
The crystal structure of human glycolate oxidase in complex with an inhibitor is described. A comparison with complexes with other inhibitors and with structures of homologous enzymes is given.
This article describes the trimeric structure of the first PDZ domain of human PSD-93 at 2 Å resolution.
PDB reference: PSD-93 PDZ1 domain, 2wl7, r2wl7sf
The structure of the extracellular domain of human CD69 was crystallized using a novel polymer precipitant: di[poly(ethylene glycol)] adipate. The structure was refined at 1.37 Å resolution.
PDB reference: extracellular domain of human CD69, 3hup, r3hupsf
crystallization communications
The cloning, expression, purification and crystallization of the mouse Elf3 C-terminal DNA-binding domain in complex with mouse type II TGF-β receptor promoter DNA are reported. The crystals were characterized and an X-ray diffraction data set was collected to a resolution of 2.2 Å.
The carbohydrate-recognition domain of the SIGN-R1 receptor from M. musculus has been crystallized by the hanging-drop vapour-diffusion method. A native data set has been collected to 1.87 Å resolution.
Open  access
access
 access
accessA 16 kDa buckwheat protein (BWp16) is a major allergen responsible for immediate hypersensitivity reactions including anaphylaxis. An immunologically active mutant of BWp16 was prepared and a three-wavelength MAD data set was collected from a crystal of selenomethionine-labelled mutant protein.
The crystallization of an HsdM subunit, a component of the methyltransferase of a putative type I restriction enzyme, from V. vulnificus YJ016 and the collection of diffraction data to 1.86 Å resolution are reported.
Selenomethionyl exo-β-1,3-galactanase from P. chrysosporium K-3 produced in Pichia pastoris was crystallized. The crystals diffracted to a resolution of 1.8 Å and belonged to space group P21.
The taming of small heat-shock proteins: crystallization of the α-crystallin domain from human Hsp27
A procedure for obtaining diffraction-quality crystals of the α-crystallin domain from human small heat-shock protein 27 with the help of limited proteolysis and rational surface mutagenesis is described.
A PCNA2−PCNA3 complex which has recently been identified from S. tokodaii strain 7 was overexpressed, purified and crystallized in two crystal forms.
Native and selenomethionine-labelled FAD synthetase from C. ammoniagenes have been crystallized by the hanging-drop vapour-diffusion method. A MAD data set for SeMet-labelled FAD synthetase was collected to 2.42 Å resolution, while data sets were collected to 1.95 Å resolution for the native crystals.
Purine nucleoside phosphorylase (PNP), which is a pivotal enzyme in the nucleotide-salvage pathway, has been expressed in Escherichia coli strain BL21 (DE3) in a soluble form at a high level. After purification of the PNP enzyme, the protein was crystallized using the sitting-drop vapour-diffusion technique.
A monomeric mutant of Azami-Green from G. fascicularis was expressed, purified and crystallized using the sitting-drop vapour-diffusion method. The crystal belonged to space group P1 and diffracted X-rays to 2.20 Å resolution.
A new crystal form of N. europaea hydroxylamine oxidoreductase (space group P21212) diffracted to 2.25 Å resolution at a third-generation synchrotron X-ray source.
Neoagarobiose hydrolase from the marine bacterium Saccharophagus degradans 2-40 was overexpressed in Escherichia coli and crystallized in the monoclinic space group C2, with unit-cell parameters a = 129.83, b = 76.81, c = 90.11 Å, β = 101.86°.
The crystallization and initial X-ray diffraction of REV7 in complex with REV3 is reported.
The crystallization and preliminary X-ray crystallographic analysis of blood coagulation factor V-activating proteinase are reported. The best crystal diffracted to 1.9 Å resolution.
The enzyme glutamine synthetase from M. truncatula has been expressed, purified and crystallized. The crystals belonged to the monoclinic space group P21 and diffracted to 2.35 Å resolution.
Purification and preliminary X-ray crystallographic analysis of maleylacetate reductase encoded by the pnpD gene is reported.
The overexpression, purification and crystallization of endonuclease IV from T. maritima are reported. The crystals belonged to the hexagonal space group P61 and diffracted to 2.36 Å resolution.


 journal menu
journal menu































![[publBio]](/logos/publbio.gif)





