issue contents
May 2015 issue
Molecular parasitology - advances in biology and supporting drug discovery

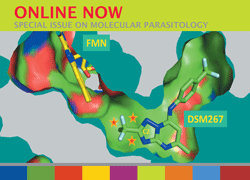
Cover illustration: Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase in complex with DSM267 and FMN. Exploiting the hydrophobic `tunnel' to the FMN binding site enabled the structure-guided design of a potent and selective inhibitor of P. falciparum DHODH. For more details see the overview of the role of three-dimensional structures in the design of therapeutics targeting parasitic protozoa in this issue (Hol, p. 485).
molecular parasitology

 access
access access
access access
access access
access access
access

 journal menu
journal menu















![[publBio]](/logos/publbio.gif)




