issue contents
November 2016 issue
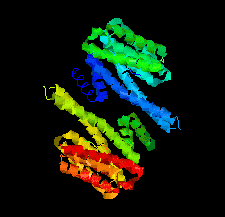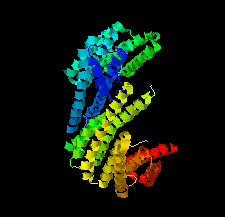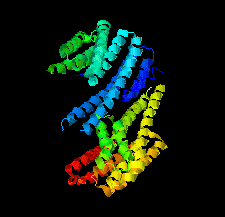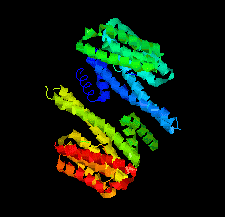
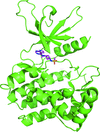
Cover illustration: Crystal structure of a yeast 14-3-3 protein from Lachancea thermotolerans bound to a human lipid kinase PI4KB-derived peptide (Eisenreichova et al., p. 799).
research communications
Crystal structures of a 14-3-3 protein (Bmh1) from the yeast L. thermotolerans in the unliganded form and in complex with a peptide from a human homologue (PI4KB) of its binding partner Pik1 reveal the high evolutionary conservation of 14-3-3 proteins. Indeed, the yeast 14-3-3 protein binds its human-derived ligand in a manner highly similar to that of human 14-3-3 proteins.
The crystal structure of a phosphoribosyl anthranilate isomerase from the hyperthermophilic archaeon T. kodakaraensis was determined in space groups P1 and C2 at 1.75 and 1.85 Å resolution, respectively.
The crystal structure of a triazolylthioacetamide inhibitor in complex with Verona integron-encoded metallo-β-lactamase 2 (VIM-2) is reported at a resolution of 1.50 Å. The structure shows that the inhibitor binds to the active site of the enzyme and reveals detailed information on the inhibitor interactions.
Open  access
access
 access
accessThe structure–affinity relationship of meditope–cetuximab complexes is investigated by measuring their affinity using surface plasmon resonance and determining their structures using X-ray crystallography.
PDB references: cetuximab Fab, complex with CQYDLSTRRLKC meditope, 5t1m; with CQHDLSTRRLKC meditope, 5euk; with GQQDLSTRRLKG meditope, 5i2i; with GQ(2-Br-F)DLSTRRLKG meditope, 5itf; with GQ(3-Br-F)-DLSTRRLKG meditope, 5ir1; with GQ(4-Br-F)DLSTRRLKG meditope, 5iop; with CQA(Ph)2DLSTRRLKC meditope, 5t1l; with CQFDYSTRRLKC meditope, 5f88; with CQFDESTRRLKC meditope, 5etu; with CQFDQSTRRLKC meditope, 5th2; with CQFDA(Ph)2STRRLKC meditope, 5t1k; with GQFDLST(Cit)RLKG meditope, 5ivz; with GQFDLSTR(Cit)LKG meditope, 5iv2; with CQFDLSTRRQKC meditope, 5ff6
The 1.34 Å resolution crystal structure of fuculose aldolase from the psychrophilic yeast G. antarctica reveals characteristics of the enzymatic site and tolerance to cold temperature.
PDB reference: fuculose aldolase, 4xxf
Active JAK1 kinase domain was crystallized with MgADP and soaked in the presence of EDTA to allow displacement of the nucleotide by compounds of interest. This high-throughput method of structure determination may be widely applicable to other protein kinases and small-molecule ATP-competitive ligands.
B. japonicum D-sorbitol dehydrogenase is NADH-dependent. It is of interest for industrial applications because of its ability to also oxidize L-glucitol and its thermostability. Here, its structure is presented and discussed.
PDB reference: D-sorbitol dehydrogenase, 5jo9


 journal menu
journal menu













![[publBio]](/logos/publbio.gif)