issue contents
December 2017 issue
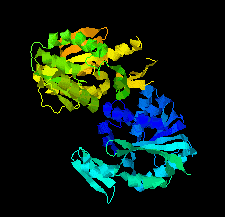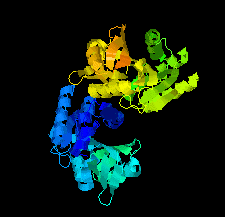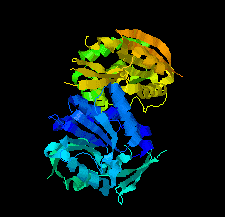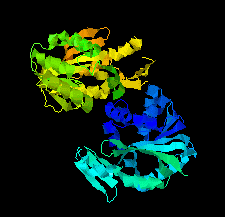
Cover illustration: SAM-dependent methyltransferase from Pyrococcus horikoshii in complex with the cofactor SAM (Pampa et al., p. 706).
research communications
The crystal structure of dTDP-4-dehydro-β-L-rhamnose reductase (RfbD) from Bacillus anthracis was determined at 2.65 Å resolution in complex with NADP+. RfbD is the fourth enzyme of the dTDP-β-L-rhamnose-biosynthetic pathway and provides evidence that RfbD homologs in Gram-positive bacteria exhibit key differences in the residues important for oligomerization and metal utilization compared with those from many Gram-negative bacteria.
PDB reference: RfbD, 3sc6
The crystal structure of a pyridoxal 5′-phosphate-dependent aspartate racemase derived from Scapharca broughtonii has been resolved at 1.90 Å resolution.
PDB reference: aspartate racemase, 5ybw
Open  access
access
 access
accessA very low cost method for carrying out imaging of trace fluorescently labeled protein crystallization plates is described.
The crystal structure of dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase (RfbC) from B. anthracis was determined at 1.63 Å resolution in complex with dTDP and pyrophosphate. RfbC is the third enzyme of the dTDP-L-rhamnose-biosynthetic pathway and only crystallized in the presence of the other three members of the pathway RfbA, RfbB and RfbD.
PDB reference: RfbC, 3ryk
The acetoacetate decarboxylase-like superfamily contains a subset of proteins that are predicted to be involved in secondary metabolism based on gene context. Swit_4259, a protein of unknown function, belongs to this emerging family V subset, in which the overall fold resembles prototypical acetoacetate decarboxylases, but the active-site architecture leads to a divergence in function.
PDB reference: Swit_4259, 5upb
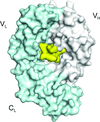
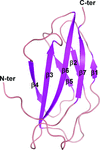
The crystal structure of an anti-idiotype VLR, VLR39, at 1.5 Å resolution is presented and compared with those of other protein-specific VLRs. Binding studies with purified components confirms a VLR39–Fv interaction.
PDB reference: VLR39, 5wk4
Open  access
access
 access
accessThe structure–affinity relationship of complexes of the cetuximab Fab with meditope peptides modified at Arg8 is investigated.
PDB references: cetuximab with aminoheptanoic acid-linked n-butylarginine meditope variant, 6au5; with aminoheptanoic acid-linked N-(3-aminopropyl)-L-arginine meditope variant, 6ayn; with aminoheptanoic acid-linked N-(3-hydroxypropyl)-L-arginine meditope variant, 6azk; with aminoheptanoic acid-linked N-carboxyethylarginine meditope variant, 6azl; with aminoheptanoic acid-linked n-octylarginine meditope variant, 6axp
The crystal structure of a fibronectin type III domain from human collagen α1 type XX (residues Pro386–Pro466) was solved at 2.5 Å resolution.
The sensor domain of the PscF chemoreceptor from the plant pathogen P. syringae pv. actinidiae has been cloned, expressed, purified and crystallized. Preliminary X-ray diffraction data from native and selenomethionine-labelled protein crystals were collected to 1.46 and 2.46 Å resolution, respectively.
The crystal structure of SAM-dependent methyltransferase from P. horikoshii in complex with the cofactor SAM was solved by X-ray diffraction. The monomeric structure consists of a Rossmann-like fold (domain I) and a substrate-binding domain (domain II). The cofactor binds at the interface between adjacent subunits.
PDB reference: SAM-dependent methyltransferase, 1ve3
addenda and errata
Free 

A corrigendum to the paper by Wu et al. [(2017), Acta Cryst. F73, 393–397] is published.


 journal menu
journal menu

















![[publBio]](/logos/publbio.gif)