issue contents
March 2019 issue
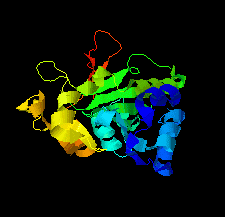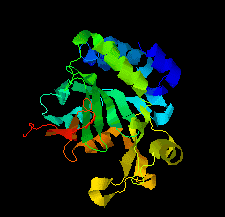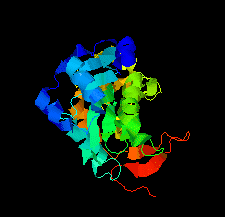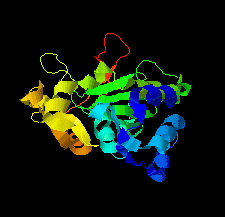
Cover illustration: Structure of the type VI immunity protein Tdi1 (Atu4351) from Agrobacterium tumefaciens [Shi et al. (2019), Acta Cryst. F75, 153-158].
editorial
Free 

Mark van Raaij welcomes our new Section Editor Janet Newman.
research communications
Further refinement of three methylenetetrahydrofolate dehydrogenase/cyclohydrolase–antifolate inhibitor complexes in the Protein Data Bank has produced models that allow a critical reassessment of ligand placement. One complex has a well ordered ligand in a catalytic site and the model provides an improved description of enzyme–inhibitor interactions. One ligand may adopt two conformations in the binding site rather than the single conformation described previously. There is no evidence to support the incorporation of the third compound in the model. Interpretation of the data supports a correlation between the models and the inhibition activity for two of the compounds. In the case of the third, inconsistencies were noted that would need to be addressed by further work.
The crystal structure of Tdi1 (Atu4351) from Agrobacterium tumefaciens is composed of a GAD-like domain and an inserted DUF1851 domain.
PDB reference: Atu4351 from Agrobacterium tumefaciens, 6itw
Osm1, a soluble fumarate reductase from Saccharomyces cerevisiae, was purified and crystallized. The crystals were found to belong to space group P21, with unit-cell parameters a = 43.88, b = 109.32, c = 49.93 Å, β = 112.25°. Crystals were obtained at 293 K and diffracted to a resolution of 1.75 Å.
X-ray crystal structures of carbonic anhydrase II (CA II) in complex with nicotinic acid and ferulic acid are presented. Previously deposited structures of CA II in complex with carboxylic acid-based compounds are compared and the general mechanisms of CA inhibition are discussed.
Large crystals of the Fenna–Matthews–Olson protein from Prosthecochloris aestuarii were grown in a new H3 space group that permitted room-temperature neutron diffraction data collection to 2.2 Å resolution.
PDB reference: Fenna–Matthews–Olson antenna complex, 6mez
The crystal structure of the heat-stable glycerol dehydrogenase from E. coli was solved at 2.79 Å resolution and compared with those of glycerol dehydrogenases from other organisms.
PDB reference: glycerol dehydrogenase from Escherichia coli, 5zxl
The reported crystallization cocktails from the REMARK 280 fields of Protein Data Bank entries are compared for similar proteins. No correlation was detected between proteins of similar (but not identical) sequence and their crystallization cocktails, suggesting that methods of determining screens based on this assumption are unlikely to result in screens that are better than those currently in use.
Heterotrimeric glutamine amidotransferase CAB (GatCAB) possesses an ammonia-self-sufficient mechanism in which ammonia is produced and used in the inner complex by GatA and GatB, respectively. Here, GatCAB crystals were successfully grown to a volume of ∼1.8 mm3, neutron diffraction data were collected and the neutron structure was solved.
The structure of the legume lectin-like domain of an ERGIC-53-like protein from E. histolytica is reported. ERGIC-53-like proteins are type I membrane proteins that belong to the class of intracellular cargo receptors and are known to be indispensable for the intracellular transport of glycoproteins.
The domain of unknown function protein CgnJ from the crocagin-biosynthetic gene cluster has no sequence homologues in the Protein Data Bank, but is a structural homologue of ComJ. CgnJ appears to bind the natural product crocagin with weak affinity. The structural homology may imply a function for CgnJ.
PDB reference: CgnJ, 6i86


 journal menu
journal menu
















![[publBio]](/logos/publbio.gif)