issue contents
September 2020 issue
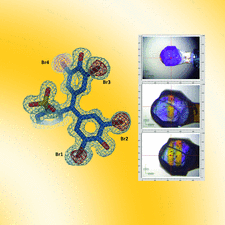
Cover illustration: Radiation damage in bromophenol blue molecules [Plaza-Garrido et al. (2020), Acta Cryst. D76, 844-855]. The Fo - Fc map shows large red blobs over the positions of the Br atoms because of the loss of these atoms from radiation damage during data collection. The right panel shows lysozyme crystals soaked at a low BPB concentration after data collection: the upper image is a crystal without irradiation and the lower image is a crystal irradiated twice at two different positions. Yellow lines appear at the irradiated positions.
scientific commentaries
Open  access
access
 access
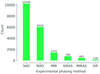
accessA new approach to in situ experimental phasing introduced by Lawrence et al. (2020, Acta Cryst. D76, 790–801) will be helpful for the macromolecular crystallography community.
CCP4
Open  access
access
 access
accessThe dnatco.datmos.org web server performs an assignment of nucleic acid conformations and presents the results for the intuitive annotation, validation, modeling and refinement of nucleic acids.
Open  access
access
 access
accessThis study shows the usefulness of integrating automated crystallographic model-building pipelines. We ran the four most used pipelines (ARP/wARP, Buccaneer, Phenix AutoBuild and SHELXE) alone and in pairwise combinations, and compared the structures that they produced based on structure completeness and Rfree.
ISDSB2019
Engrailed homeodomain (EHD) is a low-molecular-weight DNA-binding protein that can potentially act as a molecular tool to target desired DNA sites in cells. The crystal structure of tandemly connected EHDs with R53A mutations revealed that the mutation does not interfere with the important base-specific interactions of the wild-type protein; instead, it works to properly modulate the DNA-binding affinity, leading to precise recognition of the target site.
PDB reference: (EHD[R53A])2–DNA complex, 6m3d
X-ray crystal structures were determined of the wild type and an inactive mutant of α-amylase from the earthworm Eisenia fetida. Structural analyses reveal the molecular properties that are responsible for catalytic activity at low temperatures and substrate recognition.
research papers
Crystallographic structures of lysozyme in complex with the dye bromophenol blue are reported.
The low-resolution 3D structure of a plant nucleotide pyrophosphatase/phosphodiesterase (NPP) solved by small-angle X-ray scattering is reported. The general structural features of plant NPPs are inferred. Structural analogies with, and differences from, mammalian NPPs are discussed.
Open  access
access
 access
accessThe crystal structure of an epoxide hydrolase from Streptomyces sp. CBMAI 2042 was determined and revealed the conserved overall fold found in other α/β-hydrolases, despite its unusual hexameric quaternary structure. Although its primary sequence is similar to that of a Mycobacterium tuberculosis epoxide hydrolase, its active-site architecture, and particularly its volume, closely resembles the human epoxide hydrolase.
PDB reference: B1EPH2, 6unw
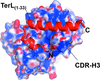
The crystal structure of Fab4 bound to the epitope of the bacteriophage P22 large terminase subunit, together with a 1.15 Å resolution crystal structure of the unliganded Fab4, which is the highest resolution ever achieved for a Fab, elucidate the principles governing the recognition of this novel helical epitope.
Open  access
access
 access
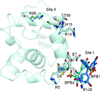
accessStructures of the cancer target human ENPP1 have been determined in the apo form, bound to nucleotides and bound to known inhibitors of the enzyme.
The crystal structure of bacteriophage T4 Spackle as determined by sulfur SAD phasing reveals a monomeric protein with a helical bundle fold and a disc-like overall shape. Spackle has an intramolecular disulfide bond and a bipolar surface-charge distribution, which are consistent with its proposed role as a lysozyme inhibitor that functions in the Escherichia coli periplasm.
PDB reference: bacteriophage T4 Spackle, 6x6o
addenda and errata
Open  access
access
 access
access

 journal menu
journal menu