issue contents
November 2008 issue
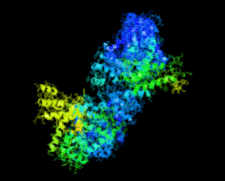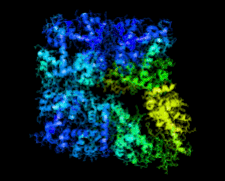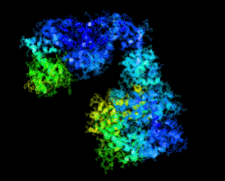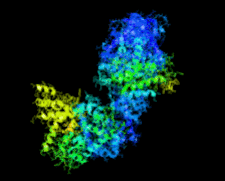
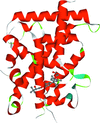
Cover illustration: Crystal structure of the Zn2+-bound form of des3-23ALG-2 (Suzuki et al., p. 974).
protein structure communications
The crystal structure of the ligand-binding domain of rat VDR in complex with a nonsecosteroidal vitamin D3 analogue YR301 has been determined at 2.0 Å resolution.
Ca2+-bound and Zn2+-bound forms of ALG-2 were crystallized and X-ray diffraction data were collected to 3.1 and 3.3 Å resolution, respectively.
The structure of Rv0802c was determined in an unliganded form to 2.0 Å resolution utilizing single-wavelength anomalous dispersion from a samarium soak that resulted in a single bound Sm3+:citrate2 complex.
The crystal structure of the MH2 domain of Mad, which closely matches those of other Smad MH2 domains, is reported at 3.2 Å resolution.
PDB reference: Drosophila Mad MH2 domain, 3dit, r3ditsf
crystallization communications
This paper describes how four general optimization techniques, growth-rate modulation, fine screening, seeding and additive screening, have been adapted for automation in a medium-throughput crystallization service facility.
2-Keto-3-deoxy-6-phosphogluconate aldolase from pathogenic S. suis, an attractive drug target, was cloned, overexpressed, purified and crystallized.
Crystals of NovN, an O-carbamoyltransferase from S. spheroides, were obtained in monoclinic and orthorhombic forms and native X-ray data were recorded to a maximum of 2.3 Å resolution.
In order to determine the protonation states of the residues within the active site of an HIV-1 protease–inhibitor complex, a crystal of HIV-1 protease complexed with inhibitor (KNI-272) was grown to a size of 1.4 mm3 for neutron diffraction study. The crystal diffracted to 2.3 Å resolution with sufficient quality for further structure determination.
Exo-α-1,5-L-arabinofuranosidase from S. avermitilis NBRC14893 was crystallized by the sitting-drop vapour-diffusion method. The crystals diffracted to a resolution of 2.2 Å and belonged to space group P212121.
The crystallization of adenylylsulfate reductase isolated from the sulfate-reducing bacterium D. vulgaris Miyazaki F is described. The crystals diffracted to 1.7 Å resolution.
The dihydropteridine reductase from D. discoideum has been crystallized. Diffraction data were collected from a rectangular-shaped crystal to 2.16 Å resolution.
Xoo2316 from X. oryzae pv. oryzae, a predicted 6-phosphogluconolactonase, the second enzyme of the pentose phosphate pathway, has been cloned, purified and crystallized. A native data set was collected to 2.4 Å resolution and a MAD data set was collected to 2.5 Å resolution.
Crystals of the nucleocapsid protein of human respiratory syncytial virus have been obtained in two forms, the better of which diffracted to 3.6 Å resolution and contained ten subunits in the asymmetric unit.
The crystallization and preliminary X-ray analysis of isomaltase is reported.
Open  access
access
 access
accessThe Fab fragment of a monoclonal anti-prostaglandin E2 antibody was prepared and its complex with prostaglandin E2 was crystallized.
Peptide deformylase (PDF) catalyzes the removal of the N-formyl group from the N-terminus of nascent polypeptides. This process is essential for the survival of bacteria. As it is an antibacterial drug target, PDF from X. oryzae pv. oryzae was cloned, purified and crystallized for preliminary X-ray studies.
Crystals of bacteriophage P1 Doc were grown in the free state, in complex with a 22-amino-acid C-terminal peptide of Phd and in complex with full-length Phd.
The membrane-targeting domains of the Rac-specific guanine nucleotide-exchange factors Tiam1 and Tiam2 were purified and crystallized.
BTL2, a thermostable enantioselective biocatalyst of interest for industrial applications, has been crystallized using the sitting-drop vapour-diffusion method. Preliminary X-ray data to 2.2 Å resolution allowed the determination of its unit-cell parameters and space group and initiation of its structure determination.
The coiled-coil domain of Atg16 was expressed, purified and crystallized. The resultant crystal diffracted to 2.5 Å resolution.
PAT, an acetyltransferase from the archaeon S. solfataricus that specifically acetylates the chromatin protein Alba, was expressed, purified and crystallized.
AcsD, an NRPS-independent siderophore synthetase, has been crystallized.
The Whirly domain of StWhy2 was crystallized both in the free form and in complex with single-stranded DNA.
Arrowhead protease inhibitor A, a member of the serine protease inhibitor family, from S. sagittifolia has been cloned, expressed, purified and crystallized in complex with bovine trypsin.
The ecto-enzymatic domain of rat E-NTPDase1 CD39 was expressed and purified and diffraction-quality crystals of the enzyme were obtained.
The plant enzyme pavine N-methyltransferase from T. flavum has been produced in E. coli, purified and crystallized and its structure has been solved.
The human Gadd45γ protein has been crystallized as a prelude towards determination of its three-dimensional structure by X-ray crystallography.
Adeno-associated virus type 6, a human DNA virus that is being developed as a vector for gene therapy, has been crystallized in a form suitable for structure determination at about 3.2 Å resolution.
The NKX2.5 homeodomain has been crystallized in complex with DNA. Diffraction data were collected to 1.7 Å resolution.
MTH909, the M. thermautotrophicus orthologue of S. cerevisiae TAN1, has been overexpressed, purified and crystallized. X-ray data were collected to 2.85 Å resolution from a crystal belonging to space group P6122 (or P6522).
The putative enolase MJ0232 from M. jannaschii was purified and crystallized. A native data set was collected to 1.85 Å resolution. Preliminary crystallographic analysis and a dynamic light-scattering experiment suggested biological relevance of the octameric state of MJ0232 in solution.


 journal menu
journal menu












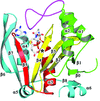























![[publBio]](/logos/publbio.gif)





