issue contents
January 2009 issue

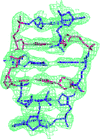
Cover illustration: A photomontage of crystal micrographs from papers published in Acta Cryst. F in 2008. Top row: the peroxiredoxin domain of a larger natural hybrid protein from Thermotoga maritima (Barbey et al., pp. 29-31); adeno-associated virus serotype 6 (Xie et al., pp. 1074-1078); and the bifunctional enzyme proline utilization A (PutA) from Bradyrhizobium japonicum (Schuermann et al., pp. 949-953). Middle row: S-adenosyl-L-homocysteine hydrolase from Lupinus luteus in complex with adenosine (Brzezinski et al., pp. 671-673); mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus (Sá-Moura et al., pp. 760-763); and flavodoxin NifF from the photosynthetic bacterium Rhodobacter capsulatus (Pérez-Dorado et al., pp. 375-377). Bottom row: polyketide synthase-1 (PKS-1) from Cannabis sativa (Taguchi et al., pp. 217-220); the MtxX protein from Methanococcus jannaschii (Dong Hae Shin, pp. 300-303); and full-length yeast tropomyosin 2 from Saccharomyces cerevisiae (Meshcheryakov et al., pp. 528-530).
editorial
structural genomics communications
The crystal structure of a phosphate-dependent exoribonuclease from B. anthracis has been solved to 1.7 Å resolution. The refined model reveals a hexameric ring structure and five bound sulfate ions per subunit. Each subunit features two sulfate ions located in the active site.
nucleic acid structure communications
The crystal structure of d(CACACG)·d(CGTGTG) was solved to a resolution of 2.05 Å in space group P21.
crystallization communications
Crystals of the complex between the Fab fragment of a human anti-interferon α therapeutic antibody and human interferon α-2A have been obtained and diffracted to 3.0 Å resolution.
A C-terminal fragment of Salmonella FlgJ, FlgJ120–316, which has peptidoglycan-hydrolysing activity, has been overproduced, purified and crystallized and the crystals have been characterized by X-ray diffraction.
Native and selenomethionine-derivatized crystals of full-length human GCIP/HHM protein were obtained. The crystals belonged to space group P3221 and the best native crystal diffracted to 3.5 Å resolution.
The native oxygen-carrier haemoglobins complex (HbII–III) is composed of haemoglobin II (HbII) and haemoglobin III (HbIII), which are found in the ctenidia tissue of the bivalve mollusc Lucina pectinata. This protein complex was isolated and purified from its natural source and crystallized using the vapour-diffusion and capillary counter-diffusion methods.
Recombinant phosphoglycolate phosphatase from S. flexneri was overexpressed, purified, characterized and crystallized using the hanging-drop vapour-diffusion method. SeMet-labelled protein was also prepared and was crystallized for phase determination using the MAD technique.
The enzyme cgHle from C. glutamicum, which has acetyl ester hydrolase activity, was crystallized in four different crystal forms. X-ray diffraction data have been collected to a resolution of 1.2 Å.
Cytosolic Trx1 containing a C33S mutant was overexpressed, purified, glutathionylated and crystallized. The crystals diffracted to 1.80 Å resolution.
The major hazelnut allergen Cor a 9 was purified from the natural source and crystallized. Diffraction data were collected to 1.9 Å resolution using a synchrotron-radiation source.
FliJ is one of the essential cytoplasmic components of the flagellar type III protein-export apparatus; it has been expressed, produced and crystallized and the crystals have been characterized by X-ray diffraction.
Glutamyl-tRNA synthetase (GluRS) is an essential enzyme for protein translation and has been considered as a target for drug development. In this paper, the cloning, expression and purification of the GluRS protein encoded by the gltX gene (Xoo1504) and its preliminarily crystallographic analysis are reported.
Crystallization conditions and preliminary X-ray diffraction analysis of the S. pneumoniae-derived pilus-associated protein sortase C are reported.
The tRNAGly acceptor-stem microhelix isoacceptor from human cytoplasm was crystallized and X-ray diffraction analysis revealed diffraction to 1.18 Å resolution. The sequence of the microhelix was derived from the gene sequence with tRNA Database ID DG9990.
international union of crystallography


 access
access


 journal menu
journal menu












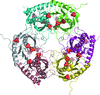












![[publBio]](/logos/publbio.gif)





