issue contents
April 2010 issue
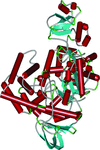
Cover illustration: Perdeuterated diisopropyl fluorophosphatase (Blum et al., p. 379).
structural communications
The structure of the rhizavidin–biotin complex was determined via combined molecular replacement approaches. Both the apo and biotin complexed structures pack in unique oligomeric states in the corresponding crystals.
The crystal structure of perdeuterated diisopropyl fluorophosphatase is reported and compared with the hydrogenated structure. Diffraction guidelines for neutron crystallography experiments are summarized.
PDB reference: perdeuterated diisopropyl fluorophosphatase, 3kgg
The three-dimensional structure of carbamate kinase from the human parasite G. lamblia has been determined at 3 Å resolution. Steady-state kinetic parameters have been determined and dsRNA-silencing experiments indicate that the enzyme is essential for the survival of G. lamblia.
PDB reference: carbamate kinase, 3kzf
Sulfur is an essential component for the biosynthesis of the sulfur-containing amino acids L-methionine and L-cysteine. The crystal structure of the periplasmic aliphatic sulfonate-binding protein SsuA from E. coli has been solved at 1.75 Å resolution in a substrate-free state. Comparison with the substrate-bound protein shows significant movement of one of the two domains.
PDB reference: SsuA, 2x26
The structure of ST0929 provides insight into the structural basis of the increase in the hydrolase side reaction that is observed for mutants in which a phenylalanine residue is replaced by a tyrosine residue in the subsite +1 tyrosine cluster of Sulfolobus sp.
PDB reference: hypothetical maltooligosyl trehalose synthase, 3hje
crystallization communications
The expression, purification and crystallization of a catalytic and ATP-binding domain of PhoR histidine kinase from D. radiodurans is described.
E. coli D-alanyl-D-alanine ligase has been crystallized in complex with ADP and D-alanyl-D-alanine and in complex with ATP and D-alanyl-D-alanine.
The collection of X-ray diffraction data from the aspartate aminotransferase of P. falciparum to a resolution of 2.8 Å is reported.
A novel family GH16 β-agarase from the marine bacterium Zobellia galactanivorans was expressed, purified and crystallized. Hexagonal crystals belonging to space group P3121 diffracted to 2.2 Å resolution, whereas orthorhombic crystals belonging to space group P212121 diffracted to 1.5–Å resolution.
The crystallization and preliminary X-ray crystallographic analysis of a 23 kDa GroEL1 fragment from M. tuberculosis H37Rv is reported.
Crystals of the oligomerization domain of the S. typhimurium histone-like nucleoid-structuring protein were obtained that diffracted X-rays to a resolution of at least 4.0 Å.
Very-long-chain acyl-CoA dehydrogenase from Caenorhabditis elegans (cVLCAD) has been crystallized in space group C2 and its X-ray diffraction data set has been collected to 1.6 Å resolution. Unlike other VLCADs that were reported to form dimers, the purified cVLCAD was found as a homotetrameric protein according to static light-scattering measurements.
The flavin-dependent enzyme FerB from P. denitrificans has been purified and both native and SeMet-substituted FerB have been crystallized. The two variants crystallized in two different crystallographic forms belonging to the monoclinic space group P21 and the orthorhombic space group P21212, respectively. X-ray diffraction data were collected to 1.75 Å resolution for both forms.
CD8α exodomain protein, a crucial immune-system factor in rhesus macaque (M. mulatta), one of the best animal models for vaccine design, was assembled and crystallized. The full structure data will contribute to future studies of immune responses in rhesus macaques.
The GyrA subunit of DNA gyrase from C. psychrerythraea strain 34H was expressed, purified and crystallized. Diffraction data were collected to a resolution of 2.60 Å.
The inverting trehalose phosphorylase from Thermoanaerobacter sp. was crystallized and its structure was determined by molecular replacement. Its purification, crystallization and preliminary X-ray diffraction analysis are presented.
The complete pneumococcal autolysin LytC has been crystallized by the hanging-drop vapor-diffusion method. A SAD data set has been collected in-house from a Gd derivative up to 2.6 Å resolution.
The hydrogenase maturation factor HypF1 from R. eutropha H16 was successfully crystallized and data sets were collected to a maximum resolution of 1.65 Å.
Crystals of the Arabidopsis abscisic acid receptors PYL9/RCAR1, PYL5/RCAR8 and PYR1/RCAR11 have been obtained.
The self-complementary octamer sequence GTGTACAC of the fully modified altritol nucleic acid has been crystallized. Diffraction data have been collected and processed to 3.0 Å resolution.
Phenylalanine hydroxylase from D. discoideum has been crystallized. Diffraction data were collected to 2.07 Å resolution.
2-Methylcitrate synthase from S. typhimurium was cloned, overexpressed, purified and crystallized. The crystals diffracted X-rays to 2.4 Å resolution and belonged to the triclinic space group P1 (unit-cell parameters a = 92.068, b = 118.159, c = 120.659 Å, α = 60.84, β = 67.77, γ = 81.92°). Self-rotation function calculations suggested the presence of a putative decamer of protein subunits in the asymmetric unit.
The crystallization of 2-keto-3-deoxy-6-phosphogluconate aldolase from Z. mobilis ZM4, a key enzyme in ethanol production, and collection of diffraction data to 1.80 Å resolution are reported.
Equine cyanomethemoglobin has been crystallized and X-ray and neutron diffraction data have been measured. Joint X-ray–neutron refinement is under way; the structural results should help to elucidate the differences between the hemoglobin R and T states.
laboratory communications
Excitation of intrinsic fluorescence at wavelengths longer than 300 nm is effective in the detection of protein crystals in crystallization trials set up in the most commonly used hardware.


 journal menu
journal menu




























![[publBio]](/logos/publbio.gif)





