issue contents
April 2013 issue
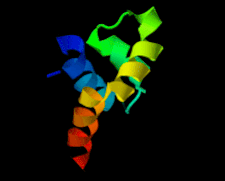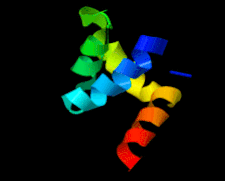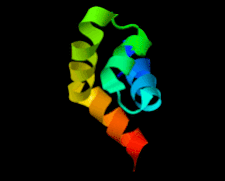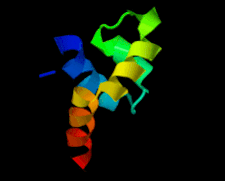
Cover illustration: The 1.58 Å resolution structure of the DNA-binding domain of bacteriophage SF6 small terminase (Benini et al., p. 376).
structural communications
Open  access
access
 access
accessThe gene encoding the hypothetical DUF1811-family protein GK0453 from G. kaustophilus was cloned and expressed. The crystal structure of the protein was determined by the molecular-replacement method and was refined to 2.2 Å resolution.
PDB reference: GK0453, 2yxy
The structural study of a bacterial anhydrolase/dipeptidase closely related to human prolidase is reported.
PDB reference: organophosphorus acid anhydrolase, 3rva
Crystallization and the structure determination at 1.6 Å resolution of LIPSBS, a thermoalkalophilic lipase from Geobacillus strain SBS-4S, are described. The crystals belonged to orthorhombic space group P212121.
PDB reference: lipase, 3auk
The structure of ligand-free glutaminyl cyclase from D. melanogaster has been determined in a novel crystal form belonging to space group I4.
PDB reference: glutaminyl cyclase, 4fwu
The crystal structure of GNA1162 from N. meningitidis has been determined at 1.89 Å resolution and its structural analysis provides important information on its function.
PDB reference: GNA1162, 4hrv
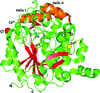
S. thermophilum produces a haem-containing catalase with phenol oxidase activity which is involved in H2O2 degradation and the oxidation of some diphenolic compounds in the absence of H2O2. The dimensions or volume of the main channel are critical in determining the rate of both movement of H2O2 into the active site and dioxygen binding, as has previously been proposed for other monofunctional catalases.
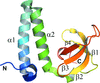
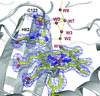
The crystal structure of the bacteriophage SF6 small terminase DNA-binding domain has been solved at 1.58 Å resolution. A model for the interaction of G1P with the DNA is proposed.
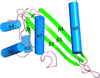
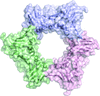
Octameric rings of DMC1 stacked into filaments in the crystal. Similar DMC1–DNA filaments have been observed previously using electron microscopy.
PDB reference: DMC1, 4hyy
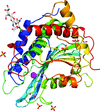
Proliferating cell nuclear antigen (PCNA) plays essential roles in DNA replication, DNA repair, cell-cycle regulation and chromatin metabolism. The PCNA from Drosophila melanogaster (DmPCNA) has been purified and crystallized.
PDB reference: PCNA, 4hk1
The deer mouse, Peromyscus maniculatus, exhibits altitude-associated variation in hemoglobin oxygen affinity. To examine the structural basis of this functional variation, the structure of the hemoglobin has been solved.
PDB reference: deer mouse hemoglobin, 4h2l
FeoB is a transmembrane protein involved in ferrous iron uptake in prokaryotic organisms. Here, the structure of the cytoplasmic domain of FeoB from Gallionella capsiferriformans is reported.
crystallization communications
SAV0479 protein from methicillin- and vancomycin-resistant S. aureus Mu50 was overexpressed and preliminary crystallographic analysis was performed. Diffraction data were collected to a resolution of 2.8 Å. Two Hiramatsu et al. (1997) references, Hiramatsu, Aritaka et al. and Hiramatsu, Hanaki et al. Please indicate in text which is meant
The tryparedoxin peroxidase from L. braziliensis was crystallized and diffraction data were collected to a maximum resolution of 3.3 Å.
The expression, purification and crystallization of the first condensation domain of viomycin synthetase are reported. An initial data set from these crystals extends to 2.9 Å resolution.
Oligomers of the haemolytic lectin CEL-III from the marine invertebrate C. echinata were crystallized and data were collected to 3.3 Å resolution using synchrotron radiation.
A complex of the tRNA-modifying enzyme 4-thiouridine synthetase and RNA was crystallized.
A DING protein from P. aeruginosa strain PA14, named PA14DING or LapC, has been crystallized. The crystals belonged to the monoclinic space group P21 and diffracted to 1.9 Å resolution.
The serine acetylxylan esterase from G. stearothermophilus (Axe2) has been crystallized in the tetragonal space group I422. Complete diffraction data sets have been measured for the selenomethionine derivative (SAD data, 1.70 Å resolution) and the wild-type enzyme (1.85 Å resolution) to be used for a full three-dimensional structural analysis of the Axe2 protein.
The CARD domain of human CARMA1 was crystallized. The crystals belonged to space group P212121, with unit-cell parameters a = 45.73, b = 53.37, c = 91.89 Å. The crystals were obtained at 293 K and diffracted to a resolution of 3.2 Å.
The Athe_0614 protein secreted by the anaerobic, extremely thermophilic and cellulolytic bacterium C. bescii was crystallized. A diffraction data set was collected to 2.7 Å resolution.
The putative ligand-binding region of serine glutamate repeat A (SgrA) protein from E. faecium was overexpressed, purified and crystallized, and its preliminary X-ray diffraction analysis is reported at 3.3 Å resolution.
HisC from M. tuberculosis was overexpressed in M. smegmatis, purified and crystallized, and its structure was solved.
The kinase domain of SAD-1 from C. elegans has been overexpressed in Escherichia coli BL21 (DE3) cells and purified to homogeneity using nickel–nitrilotriacetic acid metal-affinity, ion-exchange and gel-filtration chromatography. Diffraction-quality crystals were grown using the sitting-drop vapour-diffusion technique.
The soluble portion of TP0435, a putative periplasmic lipoprotein from the syphilis spirochete T. pallidum, has been purified and crystallized in a rhombohedral space group. A complete native data set has been collected to 2.4 Å resolution.
A recombinant form of tannase from L. plantarum was purified and crystallized. A native data set has been collected to 1.65 Å resolution.
The expression, purification, crystallization and data collection and processing of the effector protein MoHrip1 from M. oryzae are reported.
A novel effector protein was recombinantly expressed, purified and crystallized. The structure was solved using the selenium SAD method.
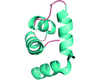
The coiled-coil domain of human PIST was crystallized at 293 K and diffracted to a resolution of 4.0 Å. The crystals were found to belong to space group P6222 or P6422, with unit-cell parameters a = b = 85.19, c = 240.09 Å, γ = 120.00°.


 journal menu
journal menu































![[publBio]](/logos/publbio.gif)





