issue contents
August 2015 issue
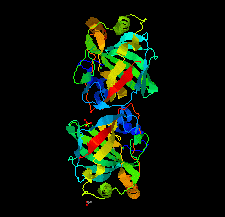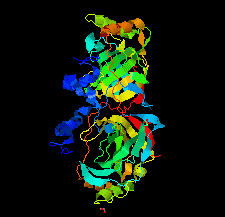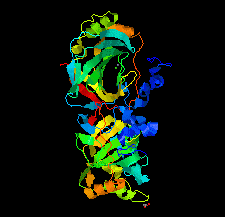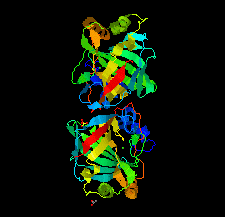
Cover illustration: ![[alpha]](/logos/entities/alpha_rmgif.gif) -Carbonic anhydrase from the human pathogen Helicobacter pylori (Compostella et al., p. 1005).
-Carbonic anhydrase from the human pathogen Helicobacter pylori (Compostella et al., p. 1005).
IYCr crystallization series
In vivo formation of protein crystals and their isolation and structural analysis by novel highly brilliant XFEL and synchrotron-radiation sources is described.
research communications
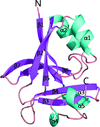
The crystal structure of human collapsin response mediator protein 1 has been determined at 3 Å resolution.
PDB reference: collapsin response mediator protein 1, 4b3z
Crystallographic X-ray diffraction studies of M. tuberculosis σJ are reported. M. tuberculosis σJ is an extracytoplasmic function σ factor that governs the cellular response to oxidative stress.
The crystal structure of recombinant prolidase from T. sibiricus was determined by X-ray diffraction to a resolution of 2.6 Å and deposited in the Protein Data Bank.
PDB reference: prolidase, 4rgz
A putative carbohydrate-binding module from a modular family 5 glycoside hydrolase from R. flavefaciens FD-1 was purified and crystallized, and data were collected from cacodylate-derivative crystals to 1.02 and 1.29 Å resolution.
A GH43 β-xylosidase from the bacterium B. licheniformis was cloned, overexpressed in E. coli BL21(DE3) cells, purified and crystallized. The crystals belonged to the monoclinic space group C2 and diffracted to 2.49 Å resolution.
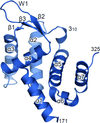
The structure of the full-length response regulator ChrA in an unphosphorylated state reveals a unique linker conformation and an interdomain reorientation.
PDB reference: ChrA, 4yn8
The halogenase HalY, which is highly similar to PltA involved in pyoluteorin biosynthesis, was expressed in E. coli and purified as a monomer. X-ray diffraction data were collected to a resolution of 1.7 Å from a crystal belonging to space group P21.
Crystals of the N-terminal domain of the nucleocapsid protein from Middle East respiratory syndrome coronavirus that diffracted X-rays to a resolution of at least 2.63 Å are described.
Open  access
access
 access
accessThe crystal structure of the human MLH1 N-terminus is reported at 2.30 Å resolution. The overall structure is described along with an analysis of two clinically important mutations.
PDB reference: human MLH1 N-terminus, 4p7a
An easy to produce, soluble variant of human glutaminyl cyclase has been validated for NMR and X-ray crystallographic screening of candidate inhibitors for lead development against neurodegenerative disorders.
The cloning, expression, purification, crystallization and crystallographic studies of ruminant-specific galectin-11 are described.
Crystallographic analysis of marine phycoerythrin is described, showing distinct sequence features.
The crystal structure of α-carbonic anhydrase, an enzyme present in the periplasm of H. pylori and a potential drug target, is described.
PDB reference: α-carbonic anhydrase from H. pylori, 4xfw
The aspartate racemase from L. sakei NBRC 15893 has been crystallized by the sitting-drop vapour-diffusion method. The crystal diffracted to 2.6 Å resolution.
The crystallization and crystallographic study of an FAD-dependent glucose dehydrogenase from A. terreus are reported.
Open  access
access
 access
accessThree crystal structures of recombinant P. aeruginosa FabF are reported: the apoenzyme, an active-site mutant and a complex with a fragment of a natural product inhibitor. The characterization provides reagents and new information to support antibacterial drug discovery.
Ectoine synthase from S. alaskensis, which catalyzes the production of the stress protectant ectoine, was crystallized, yielding crystals that diffracted to 1.0 Å resolution.
The crystallization of the enzyme NosA, which catalyzes nosiheptide maturation, is reported.
The cloning, expression and crystallographic studies of SarV from S. aureus are reported.
The crystal structure of c4763, a uropathogenic E. coli-specific protein, at a resolution of 1.45 Å reveals that it is a structural homologue of allophanate hydrolase, which is involved in the urea cycle. These results suggest that c4763 might be involved in urea utilization, which is necessary for bacterial survival in the urinary tract.
PDB reference: c4763, 5c5z
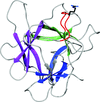
Roquin mediates mRNA degradation by recognizing the constitutive-decay element. Crystal structures of the ROQ domain of Roquin-2 were determined in ligand-free and RNA-bound forms, in which Roquin-2 recognized RNA forming a stem-loop structure.
The crystal structure of a serine protease inhibitor from B. bauhinioides was determined at 1.4 Å resolution and compared with the structures of other Kunitz-type inhibitors, including the closely related dual-specificity inhibitor BbCI.
PDB reference: BbKI, 4zot
The crystallization and X-ray diffraction analysis of RsmA from S. aureus are reported.
CoA binds in an extended conformation to the amino-terminal domain of the α-subunit of GTP-specific succinyl-CoA synthetase.
PDB reference: GTP-specific succinyl-CoA synthetase, CoA complex, 4xx0
The details of the preparation of a large crystal of X-2 L110F, a mutated carbohydrate-binding module of a bacterial xylanase, in complex with a branched xyloglucan oligosaccharide ligand are described. Details include crystallization, H/D exchange, room-temperature X-ray and neutron data collection, initial joint X-ray/neutron refinement using phenix.refine and initial structure analysis.
V. vulnificus VatD was expressed, purified and crystallized. Diffraction data were collected to 2.60 Å resolution for apo VatD and to 2.03 Å resolution for a VatD–desferrioxamine B–Fe3+ complex.
Crystallization and X-ray crystallographic studies of the archaeal ribosomal protein L11 from M. jannaschii are reported.
The high-resolution crystal structure of a cAMP-dependent protein kinase is reported.
PDB reference: cAMP-dependent protein kinase, 4wih
The structure of a eukaryotic GUN4 is compared with prokaryotic GUN4 structures.
PDB reference: GUN4, 4ykb
Open  access
access
 access
accessThe crystal structure of the protein product of the C. acetobutylicum ATCC 824 gene CA_C0359 is structurally similar to YteR, an unsaturated rhamnogalacturonyl hydrolase from B. subtilis strain 168. Substrate modeling and electrostatic studies of the active site of the structure of CA_C0359 suggests that the protein can now be considered to be part of CAZy glycoside hydrolase family 105.
PDB reference: C. acetobutylicum ATCC 824 glycoside hydrolase, 4wu0
The biochemical characterization and X-ray crystallographic analysis of branching enzymes from the cyanobacterium Cyanothece sp. ATCC 51142 are reported.


 journal menu
journal menu





























![[publBio]](/logos/publbio.gif)