issue contents
June 2016 issue
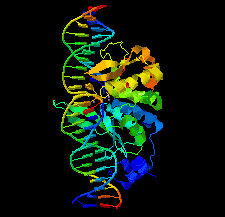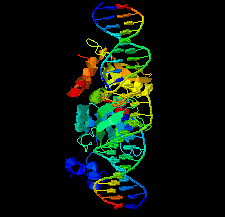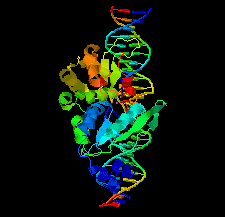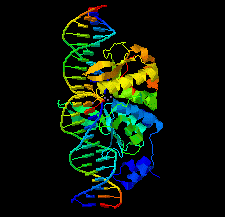
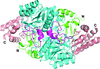
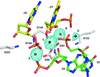
Cover illustration: Structure of the I-SceI nuclease complexed with its dsDNA target and three catalytic metal ions (Prieto et al., p. 473). Homing endonucleases such as I-SceI are highly specific DNA-cleaving enzymes that recognize and cleave long stretches of DNA. The engineering of these enzymes provides instruments for genome modification in a wide range of fields, including gene targeting.
research communications
 access
access access
access access
access

 journal menu
journal menu














![[publBio]](/logos/publbio.gif)