issue contents
August 2016 issue

Cover illustration: The human DNA-repair protein RAD52 containing surface mutations (Saotome et al., p. 598).
research communications
Open  access
access
 access
accessThe structure of the hEAG PAS domain is presented and compared with similar segments of human and insect ion channels.
PDB reference: hEAG PAS domain, 5j7e
VP24 is one of the major envelope proteins of White spot syndrome virus. In order to facilitate purification, crystallization and structure determination, the predicted N-terminal transmembrane region of approximately 26 amino acids was truncated from VP24 and several mutants were prepared to increase the ratio of methionine for subsequent data collection using the SAD method.
AKAP18γ/δ is recognized for its involvement in modulating cardiac Ca2+ release and re-uptake and is a potential drug target in cardiovascular disease. The atomic resolution crystal structure of the central domain of AKAP18γ/δ complexed with malonate presents an optimal template for structure-guided drug design.
PDB reference: AKAP18γ/δ, 5jj2
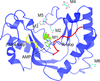
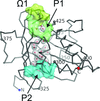
The structure of the N-terminal half of the human RAD52 protein containing three alanine substitutions was solved at 2.4 Å resolution. The structure revealed a previously unobserved interaction between RAD52 rings in which the C-terminal end of one of the monomers in the RAD52 ring associates with the positively charged groove of a symmetry-related RAD52 ring in the crystal lattice.
PDB reference: RAD521–212 K102A/K133A/E202A mutant, 5jrb
Open  access
access
 access
accessL-Arabinonate dehydratase and D-xylonate dehydratase from the IlvD/EDD family were crystallized by the vapour-diffusion method. Diffraction data sets were collected to resolutions of 2.40 and 2.66 Å from crystals of L-arabinonate dehydratase and D-xylonate dehydratase, respectively.
Open  access
access
 access
accessThe structure of a START-domain protein known to bind lutein in the human retina is reported to an improved resolution limit. Rigid-body docking demonstrates that at least a portion of lutein must protrude from the large tunnel-like cavity characteristic of this helix-grip protein and suggests a mechanism for lutein binding specificity.
PDB reference: cholesterol and lutein-binding domain of human STARD3, 5i9j
The crystal structure of the substrate-recognition domain of an Fbs1 mutant was determined at a resolution of 2.3 Å. Comparison of the wild-type and mutant Fbs1 structures provides insight into the structural features of this carbohydrate-binding site.
PDB reference: substrate-recognition domain of an Fbs1 mutant, 5b4n
Crystal structures of a subunit of the formylglycinamide ribonucleotide amidotransferase, PurS, from T. thermophilus, S. tokodaii and M. jannaschii are determined and their structural characteristics are analyzed.
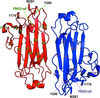
The monoclonal antibody UIC2 binds specifically to the extracellular region of the human P-glycoprotein in a conformationally sensitive manner. The crystal structure of the Fab fragment of UIC2 is reported.
PDB reference: antibody fragment of UIC2, 5jue
The crystal structure of Rv3899c184–410 was found to contain a compact helix bundle and an α/β/α sandwich folding domain.
PDB reference: Rv3899c184–410, 5imu
The interaction between PICH and BEND3 is dissected and the domains involved in their interaction have been crystallized.
The crystal structure of AibC, the terminal reductase in myxococcal de novo isovaleryl coenzyme A biosynthesis, has been determined at 2.55 Å resolution.
PDB reference: AibC, 5kis


 journal menu
journal menu











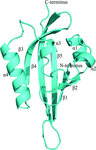



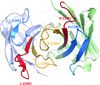


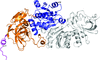
![[publBio]](/logos/publbio.gif)