issue contents
November 2017 issue
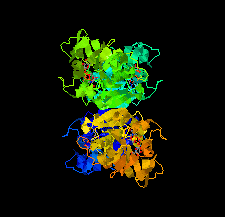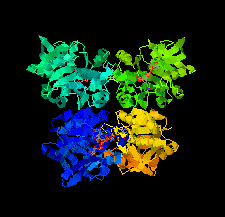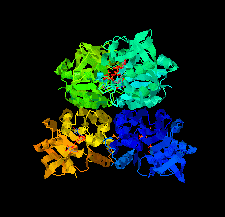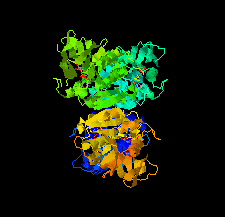
Cover illustration: Potential drug targets for anthrax: structure of dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA) from Bacillus anthracis (Baumgartner et al., p. 621).
research communications
The crystal structure of the N253F mutant of trehalose synthase from D. radiodurans reveals a new open conformation with an empty active site, which may provide a snapshot of the apoenzyme prior to substrate binding.
PDB reference: trehalose synthase, N253F mutant, 5gtv
Open  access
access
 access
accessMutations introduced into S19, a recombinant staphylococcal enterotoxin B vaccine candidate, contribute to its reduced cytokine induction in vivo by removing hydrogen bonds and allowing interaction with the T-cell receptor beta chain and major histocompatibility complex class II.
PDB reference: staphylococcal enterotoxin B mutant S19, 5xz0
The periplasmic form of dipeptidyl aminopeptidase IV from P. mexicana WO24 was crystallized in complex with the tripeptide Lys-Pro-Tyr. X-ray diffraction data were collected to 1.90 Å resolution.
PhyH-DI, a β-propeller phytase from Bacillus sp. HJB17 that interacts with PhyH-DII and other typical β-propeller phytases, can increase the efficiency of phytases in hydrolyzing phytate. Native and selenomethionine-substituted PhyH-DI were expressed, purified and crystallized.
Three mutants of B. megaterium porphobilinogen deaminase that affect a key catalytic residue (Asp82) have been analysed. Comparison with the wild-type enzyme shows significant domain movements and suggests that the enzyme adopts `open' and `closed' conformations in response to substrate binding.
The crystal structure of glucose-1-phosphate thymidylyltransferase (RfbA) from B. anthracis was determined at 2.3 Å resolution in complex with the reaction products dTDP-α-D-glucose and pyrophosphate. RfbA is the first enzyme of the dTDP-L-rhamnose pathway and does not exhibit the structural features that form the allosteric site in its closest RmlA homologs.
PDB reference: RfbA, 4ecm
Open  access
access
 access
accessThe cloning, expression, purification, crystallization and preliminary X-ray crystallographic analysis of pteridine glycosyltransferase from Chlorobium tepidum are reported.
Point mutations in the CoA-binding region of M. tuberculosis pantothenate kinase lead to structures similar to that of the E. coli enzyme through concerted movements at the dimer interface and the ligand-binding region.


 journal menu
journal menu













![[publBio]](/logos/publbio.gif)