issue contents
May 2019 issue

Cover illustration: Using cryo-EM and X-ray crystallography, the atomic resolution structure of the tetrameric form of the proton-dependent oligopeptide transporter PepTSo2 has been determined [Nagamura et al. (2019), Acta Cryst. F75, 348-358] and the novel oligomerization mechanism revealed.
editorial
Open  access
access
 access
accessThe policy of IUCr Journals on diffraction data is defined.
research communications
The structure of the haloalkane dehalogenase DpcA from the psychrophilic bacterium Psychrobacter cryohalolentis K5, an attractive enzyme for biotechnological applications, was solved at the atomic resolution of 1.05 Å. The enzyme possesses main and slot tunnels with the shortest lengths in comparison with other haloalkane dehalogenases. Structural comparisons show major differences in the region of the α4 helix of the cap domain, which is one of the determinants of the properties of the tunnels. The structural information on DpcA establishes a basis for understanding its catalytic properties and can guide the modification of this cold-adapted enzyme for various biotechnological applications.
PDB reference: DpcA from Psychrobacter cryohalolentis K5, 6f9o
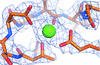
The complex of two key proteins of the SUMOylation pathway is presented to 2.6 Å. This work reveals the polar nature of the interaction between the proteins, and shows how the essential QQTGG motif of SUMO2 is arranged relative to the active site of SENP1.
PDB reference: SENP1–SUMO2 complex, 6nnq
The 3.1 Å resolution single-particle cryo-electron microscopy structure of the RNA-bound Ebola virus nucleoprotein helical filament provides molecular details of protein–protein and protein–RNA interactions.
PDB reference: Ebola virus NP–RNA helical filament, 6nut
Open  access
access
 access
accessUsing cryo-EM and X-ray crystallography, the atomic resolution structure of the tetrameric form of the proton-dependent oligopeptide transporter PepTSo2 was determined and the novel oligomerization mechanism was revealed.
A crystal structure of Mycobacterium tuberculosis filamenting temperature-sensitive mutant Z, which has elucidated a novel conformation involving the T9 loop and the nucleotide-binding pocket, is presented.
PDB reference: FtsZ from Mycobacterium tuberculosis, 5v68
New crystal structures of the N-terminal domain of LpoA from Escherichia coli and Haemophilus influenzae revealed the ability of this domain to flex between relatively flat and curved structures and to vary the width of its central groove.
A simple pipeline is introduced for de novo protein structure determination by calcium SAD phasing using an in-house diffractometer equipped with a chromium rotating anode.
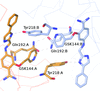
The benzamidine-derived ligand GSK144 inhibits the target enzyme kallikrein 5 (KLK5) with a pIC50 of 7.1. X-ray crystallography of KLK5 proved challenging, with the only structure solved being that with GSK144. To enable structure-guided design in support of medicinal chemistry, a KLK6-derived crystal system was developed. Here, a biochemical pharmacological assessment of the KLK6 surrogate enzymes compared with KLK5 is described and details of the X-ray structures obtained with GSK144 are discussed.
Grx1, a cytosolic thiol-disulfide oxidoreductase, actively maintains cellular redox homeostasis using glutathione substrates. Here, the crystal structure of reduced yGrx1 at 1.22 Å resolution is reported and compared with the existing structures of the oxidized and glutathionylated forms. This revealed structural differences in the conformations of residues neighbouring the Cys27–Cys30 active site, which accompany alterations in the redox status of the protein.
PDB reference: reduced Grx1, 6mws
addenda and errata
Free 

A correction to the article by Sharma et al. [(2018), Acta Cryst. F74, 656–663] is published.


 journal menu
journal menu















![[publBio]](/logos/publbio.gif)