issue contents
Room temperature biological crystallography
Edited by Roberto Steiner
This virtual issue collects together articles on room temperature biological crystallography.
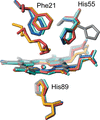
Cover illustration: Some of the room temperature structures described in this issue.
Free 

Room-temperature biological crystallography has seen a resergence in recent years and a collection of articles recently published in IUCrJ, Acta Cryst. D Structural Biology and Acta Cryst. F Structural Biology Communications, have been collected together to produce a virtual special issue at https://journals.iucr.org/special_issues/2022/RT/.
The determination of the structure of the multifunctional globin dehaloperoxidase using multiple room-temperature methods is described. Structures obtained by serial femtosecond crystallography, serial synchrotron crystallography, neutron diffraction and serial Laue crystallography are compared and two oxidation states of the enzyme are contrasted.
PDB references: ferric DHP-B, neutron diffraction structure, 7jor; X-ray structure from crystal used for neutron diffraction, 7kfm; serial femtosecond crystallography structure, 7adf; serial synchrotron crystallography structure, 7acp; serial Laue crystallography structure, 7adq; oxyferrous DHP-B, joint X-ray/neutron structure, 7kcu; serial femtosecond crystallography structure, 7adx
Open  access
access
 access
accessLipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre
Serial crystallography relies upon the growth of microcrystals at high concentration. An LCP crystallization protocol for the Blastochloris viridis photosynthetic reaction centre that utilizes seeding from detergent-grown crystals is reported.
Analysis of high-resolution data indicated that X-ray damage has little impact on conformational ensemble information from protein crystals at room temperature (277 K). In contrast, X-ray damage can alter conformational distributions obtained from cryo-cooled crystals under common experimental conditions, complicating structural interpretations in some instances. The results presented provide a strong motivation for the expanded use of room-temperature X-ray crystallography.
Download citation


Download citation


Open  access
access
 access
accessA room-temperature X-ray crystallographic method using temperature as a trigger to record movie-like structural snapshots has been developed and applied to study ligand binding and protein plasticity.
PDB references: endothiapepsin–TL00150 complex, 7qlt; 7qlu; 7qlv; 7qlw; 7qlx; 7qly; 7qlz; 7qm0; 7qm1; 7qm3; 7qm4; 7qm5; 7qm6; 7qm7; 7qm8; 7qm9; 7qma; 7qmb; 7qmc; 7qmd; 7qme; 7qmf; 7qmg; 7qmh; 7qmi; 7qmj; 7qmk; 7qml; 7qmm; 7qmn; 7qmo; 7qmp; 7qmq; 7qmr; 7qms; 7qmt; 7qmu; 7qmv; 7qmw; 7qmx; 7qmy; 7qmz; 7qn0; 7qn1; 7qn2; 7qn3; 7qn4
X-ray crystallography at variable temperature for SARS-CoV-2 Mpro reveals a complex conformational landscape, including a mobile metal at the catalytic dyad, mercurial conformational heterogeneity at various sites, and an intramolecular network bridging the active site and dimer interface.
The use of room-temperature crystallography to probe protein dynamics on the second-to-minute timescale is demonstrated by monitoring the release of a photoequilibrium induced in crystals of a LOV domain. The slow protein dynamics components include the relaxation of a photoadduct and a crystalline phase transition. The result of the latter is the formation of a non-crystallographic dimer in which the C-termini of the two monomers reorder in distinct conformations on different timescales.
The structure of the full-length Drosophila cryptochrome as solved by fixed-target serial synchrotron crystallography at the Cornell High Energy Synchrotron Source is presented. Scaling and filtering methods were performed to improve the overall quality of the final model.
PDB reference: Drosophila cryptochrome, room temperature, 7ud0
Room-temperature crystallography enables researchers to resolve the conformational heterogeneity of structures. Here, the native SAD phasing of four structures at 295 K highlights the strengths of room-temperature diffraction experiments, including detailed anomalous difference maps and alternate conformations that are well supported by the electron density.
Using the CFEL TapeDrive, minimization of sample consumption and data collection times needed for serial synchrotron crystallography (SSX) were achieved and the first room-temperature structures of two enzymes are described as well as an assessment of the benefit of SSX for a radiation-sensitive protein.
PDB references: RT SSX structures:CTX-M-14, 7zpv; CTX-M-14 (10k dataset), 8af7; K. pneumoniae CTX-M-14 (5k dataset), 8af8; N. haematococca GH11 xylanase (40k dataset), 8af4; GH11 xylanase (10k dataset), 8af5; GH11 xylanase (4k dataset), 8af6; GH11 xylanase (1k dataset), 7zq0; cofactor-free A. flavus urate oxidase complex, 7qar
Download citation


Download citation


Open  access
access
 access
accessAn in situ X-ray diffraction measurement system using a crystallization plate has been constructed at the SPring-8 protein crystallography beamline. Utilizing small-wedge measurements and incorporating a liquid dispenser to prepare protein–ligand complex crystals, this system will make ligand screening possible.
Open  access
access
 access
accessThe first serial synchrotron crystallographic structure of apo PTP1B is reported and the conformational heterogeneity at several allosteric sites is compared with that seen in prior structures.
Open  access
access
 access
accessKey physical principles and methods for collecting crystallographic data from biomolecular systems at room temperature and, more generally, at temperatures between ∼200 and ∼350 K are reviewed and discussed.
Anomalous information was obtained from model lysozyme, thaumatin and proteinase K crystals from 220 K up to physiological temperatures.
Serial femtosecond X-ray crystallography was performed using a liquid-jet system on bacterial copper amine oxidase microcrystals anaerobically premixed with an amine substrate. The structure determined at 1.94 Å resolution revealed catalytic intermediates in two reduced forms, confirming that anaerobic conditions were well maintained throughout the measurements.
PDB reference: catalytic intermediate of copper amine oxidase, 7ynh

 menu
menu