issue contents
October 2018 issue
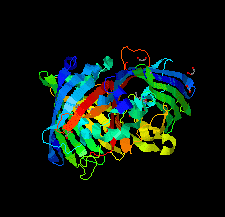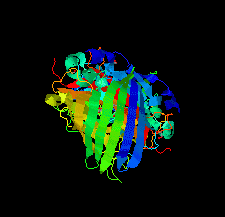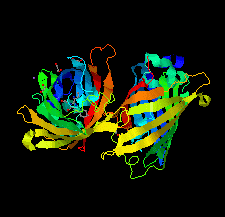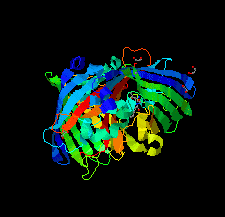
Cover illustration: The crystal structure of superfolder green fluorescent protein with Asp133 mutated to the unnatural amino acid 4-nitro-L-phenylalanine [Maurici et al. (2018), Acta Cryst. F74, 650-655].
research communications
The first cryo-neutron diffraction experiment for a DNA oligonucleotide crystal is reported. Data collection in less than two days with a crystal of approximate size 0.25 mm3 yielded a data set of >90% completeness to a resolution of 1.7 Å.
Crystal structures of apo malate dehydrogenase (MDH) from Methylobacterium extorquens, MDH bound to NAD+, and MDH with oxaloacetate and ADP-ribose revealed conformational changes, closing the active site upon coenzyme and substrate binding. In the ternary complex, His284 is in position to donate a proton in the formation of (2S)-malate.
The crystal structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae was determined at 2.1 Å resolution in complex with NAD+ and oxaloacetate. The structure of the active-site loop in MDH3 differed from those of mitochondrial and cytosolic malate dehydrogenases.
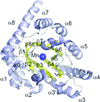
Peroxiredoxin A (PrxA) from Arabidopsis thaliana chloroplasts belongs to the 2-Cys Prx family. The crystal structure of a PrxA C119S mutant at 2.6 Å resolution shows the protein exists as a decamer, which was corroborated in solution. The structure is in the reduced state, suitable for the approach of peroxide.
PDB reference: Arabidopsis thaliana peroxiredoxin A C119S mutant, 5zte
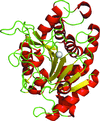
Staphylococcus aureus ribonuclease-P-protein subunit (RnpA) is a promising antimicrobial target. The crystal structure of RnpA at 2.0 Å resolution shows key similarities with RnpA structures from bacteria and archaea and will be instrumental in designing inhibitors targeting RnpA-mediated RNA processing.
PDB reference: Staphylococcus aureus ribonuclease-P-protein subunit, 6d1r
A mutant of carboxypeptidase T (CPT) has been generated in which the amino-acids of the S1′ subsite are substituted by the corresponding residues from pancreatic carboxypeptidase B (CPB). The mutant enzyme retained the broad selectivity of wild-type CPT. A comparison of the structures of CPT, the mutant and CPB showed that the S1′ subsite has not been distorted by the mutagenesis and adequately reproduces the structure of the CPB S1′ subsite, but differs from CPB in substrate selectivity.
Metallo-β-lactamases (MBLs) may hydrolyze the β-lactam rings of antibiotics. PNGM-1 is a subclass B3 deep-sea sediment MBL that predates the antibiotic era. Crystals of native and selenomethionine-substituted PNGM-1 diffracted to 2.1 and 2.3 Å resolution, respectively. They belonged to space group P21 and probably contain 6–10 molecules in the crystallographic asymmetric unit.
X-ray crystal structures of superfolder green fluorescent protein with the unnatural amino acid 4-nitro-L-phenylalanine (pNO2F) independently incorporated at two sites in the protein are reported. These structures support the use of pNO2F as a relatively nonperturbative spectroscopic reporter of local protein environment.
X-ray structures of the ETSi domain of human ERG3 and of its complex with the E74 promoter DNA sequence are reported. Comparison of the ETSi–DNA9 structure with known structures of ETS class 1 protein–DNA complexes shows the similarities and differences in the promoter DNA binding and specificity of the class 1 ETS proteins.
Open  access
access
 access
accessIn order to facilitate structure-based drug-design efforts, a set of structures of orotidine 5′-monophosphate decarboxylase from the malaria parasite P. falciparum were re-refined. This refitting greatly improves the global statistics of all three structures and shows that adding the substrate orotidine 5′-monophosphate to the enzyme (PDB entry 2za1) generates a complex with the product UMP, rather than the substrate OMP as initially interpreted.
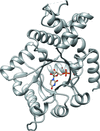
The crystal structure of A. globiformis M30 ketose 3-epimerase was determined at 1.96 Å resolution.
PDB reference: A. globiformis M30 ketose 3-epimerase, 5zfs
Open  access
access
 access
accessHuman mitochondrial manganese superoxide dismutase (MnSOD) is a major player in combating reactive oxygen species in the human body. Methods have been found to control the redox state of the active-site metal of large perdeuterated MnSOD crystals. Neutron diffraction data from these crystals were collected to study the effect of the redox state on proton location. These methods can be applied to other crystal systems where information on the location of protons in specific chemical states is needed.


 journal menu
journal menu












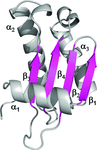



![[publBio]](/logos/publbio.gif)