issue contents
October 2021 issue
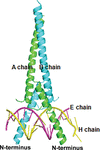
Cover illustration: The crystal structure of DynU16 [Alvarado et al. (2021), Acta Cryst. F77, 328–333]. DynU16 is a putative di-domain protein homologous to StAR-related lipid-transfer (START/Bet v1-like) domain-containing proteins and is found in the dynemicin-biosynthetic gene cluster of Micromonospora chersina.
research communications
Open  access
access
 access
accessThe crystal structure of DynU16, a protein identified in the dynemicin-biosynthetic gene cluster of Micromonospora chersina, was determined using iodide phasing and reveals a di-domain helix-grip fold.
PDB reference: DynU16, 6v04
Peptidyl-arginine deiminases (PADs) catalyse the post-translational modification of arginine residues in numerous proteins. The human type III isoform (PAD3) is specifically expressed in skin tissue and is required for normal skin and hair formation. The structure of human PAD3 has been determined at 2.8 Å resolution, giving insight into the substrate specificity of the enzyme.
PDB reference: peptidyl-arginine deiminase type III, 6ce1
TYE7 has abundant physiological functions. Here a complex of DNA with the C-terminal domain of TYE7 from Saccharomyces cerevisiae has been determined. Detailed structural analysis shows that Arg193 in TYE7 is unusual compared with Arg193 in other similar structures.
PDB reference: C-terminal domain of TYE7, complex with DNA, 7f2f
Four crystal structures of human coronavirus NL63 main protease (Mpro) in the apo form at different pH values are reported at resolutions of up to 1.78 Å. Comparison with Mpro from other human betacoronaviruses such as SARS-CoV-2 and SARS-CoV reveals common and distinct structural features in the different genera and extends knowledge of the diversity, function and evolution of coronaviruses.
methods communications
Microcrystals of bacterial copper amine oxidase were prepared by combining micro-seeding and batch crystallization, and were used to determine a radiation-damage-free, nonfrozen structure by serial femtosecond X-ray crystallography.
The cystallization and structure determination of the armadillo repeat domain of Drosophila SARM1 are described. The phase problem was solved using the MIRAS technique with autoSHARP, combining diffraction data from native, selenomethionine-labelled and bromide-soaked crystals.
PDB reference: armadillo repeat domain of Drosophila SARM1, 7lcz
Native mass spectrometry was used to screen for nanobodies that allowed crystallization of the transcription regulator PaaR2.


 journal menu
journal menu
















![[publBio]](/logos/publbio.gif)





