issue contents
May 2025 issue

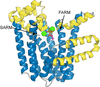
Cover illustration: The structure of the ligand-binding domain of peroxisome proliferator-activated receptor gamma in complex with piperine was determined, resolving discrepancies among predictions of their interaction using computational methods [Egawa et al. (2025), Acta Cryst. F81, 201–206].
research communications
Download citation


Download citation


High-throughput protein crystallography is used at the heart of a pipeline to accelerate the discovery of bioactive natural products by capturing hits directly from unpurified biota samples using protein crystals.
Open  access
access
 access
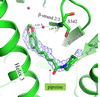
accessWe report the structure of M. tuberculosis isoprenyl diphosphate synthase Rv2173 in three forms, including two with substrate (isoprenyl diphosphate and dimethylallyl diphosphate) occupying the allylic substrate site in different binding poses, with different numbers of metal ions bound. The homodimeric structures possess a canonical all-α-helical trans-isoprenyl diphosphate synthase fold, which supports small but significant differences, notably in the ordering of the C-terminus that closes the active site.
This study reports the co-crystallization of piperine with the peroxisome proliferator-activated receptor gamma (PPARγ) ligand-binding domain, providing detailed insights into the piperine binding position and its interactions with specific amino acids within the receptor. X-ray crystallographic analysis of the co-crystal structure revealed that piperine binds in the ligand-binding pocket of PPARγ via hydrogen-bonding and hydrophobic interactions, suggesting that it plays a role as a partial agonist or antagonist and thereby holds promise as a natural alternative to synthetic PPARγ modulators.
PDB reference: PPARγ ligand-binding domain in complex with piperine, 9l3t
Download citation


Download citation


Open  access
access
 access
accessArchaeal organisms possess Cdc45 and GINS homologs that are likely to serve similar functional roles, but their structural interactions with the MCM helicase and hence their mechanism of MCM activation are not as well understood as for their eukaryotic counterparts. We present the crystal structure of the S. solfataricus GINS tetrameric complex and illustrate that a subdomain would need to move to accommodate known archaeal GINS-complex interactions and to generate an S. solfataricus CMG complex analogous to that of eukaryotes.
PDB reference: Saccharolobus solfataricus GINS tetramer, 9moj
methods communications
The crystallization and preliminary X-ray crystallographic analysis of a de novo-designed protein featuring a rare left-handed βαβ motif are reported.
addenda and errata
Free 


 journal menu
journal menu














![[publBio]](/logos/publbio.gif)





