issue contents
October 2024 issue
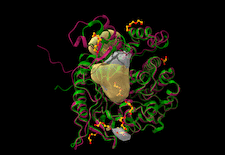
Cover illustration: β-Glucosidase from the thermophilic bacterium Caldicellulosiruptor saccharolyticus (Bgl1) [Sotiropoulou et al. (2024), Acta Cryst. D80, 733–743]. Superposition of the crystal structures of Bgl1 (red) and β-glucosidase from C. cellulovorans (green). The catalytic site of Bgl1 and an additional adjacent cavity that was detected by the DeepSite algorithm are highlighted in surface representation (yellow).
CCP4
Open  access
access
 access
accessThis article examines the application of anomalous scattering for the identification of elements within crystal structures of macromolecules.
research papers
Open  access
access
 access
accessBeamstops and beamstop shadows can lead to erroneous reflection measurements in crystallographic diffraction experiments, which is a common problem in data processing. Here, a method is presented that combines crystallographic statistics with machine learning to robustly detect such reflections.
Open  access
access
 access
accessCrystal structures of a β-glucosidase from the thermophilic bacterium C. saccharolyticus (Bgl1) and its complex with glucose were determined at 1.47 and 1.95 Å resolution, respectively. Comparison of Bgl1 with sequence or structural homologues showed that Bgl1 is quite similar except for two regions, one of which seems to be a unique insertion of residues that is only present in Bgl1.
Open  access
access
 access
accessA description is given of new deep-learning tools that analyse experimental micrographs of crystallization experiments to enable the automation of outcome classification, crystal detection and determination of locations to dispense compounds for fragment-based drug discovery at Diamond Light Source.

 journal menu
journal menu